|
|
||||||||||
|
What is Pseudomonas aeruginosa? Pseudomonas aeruginosa группа a b c по антибиотикамPseudomonas aeruginosa morphology, growth characteristics and microscopic appearance of P.aerginosa. Images of Pseudomonas aeruginosa, infections caused by Pseudomonas aeruginosa.Pseudomonas aeruginosa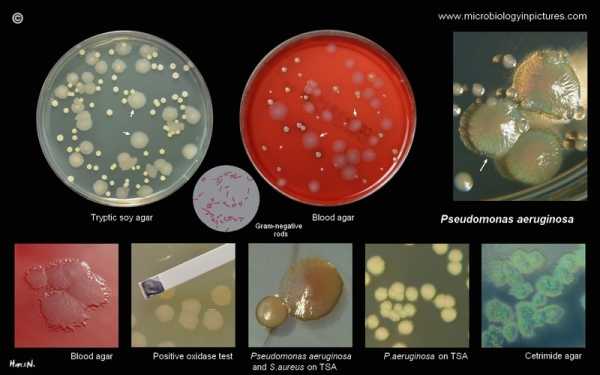 Pseudomonas aeruginosa is a Gram-negative, rod-shaped, motile organism (polar flagella) which characteristically produce water-soluble pigments which diffuse through the medium. The best known are pyocyanin (blue-green), pyoverdine (yellow-green, fluorescent), and pyorubin (red-brown, produced by a small proportion of strains). It is commonly found free living in moist environments but is also a pathogen of plants, animals, and humans. Pseudomonas aeruginosa is often preliminarily identified by its typical odor in vitro. The smell is described as grape-like, tortilla-like or "Philadelphus coronarius-like" (production of aminoacetophenone). It is not an active fermenter of carbohydrates and produces acid, but no gas, in glucose and is lactose-negative. The oxidase and catalase test for P.aeruginosa are positive. Although an aerobic atmosphere is necessary for optimal growth, most strains will multiply slowly in an anaerobic environment if nitrate is present as a hydrogen acceptor. It is able to grow at 42°C. The combination of pyocyanin production and the ability to grow at 42°C is sufficient to distinguish P.aeruginosa from other Pseudomonas spp. (e.g., P.fluorescens, P.putida, P.stutzeri, P.putrefaciens). Certain strains of P.aeruginosa may not produce pyocyanin. Pseudomonas aeruginosa is an opportunistic pathogen that infects burns, wounds, surgical incisions and sites of catheterization. It is the most common cause of infections of burn injuries and of the outer ear (otitis externa). It can, in rare circumstances, cause community-acquired pneumonias, as well as ventilator-associated pneumonias. P. aeruginosahas has immense potential to develop resistance against antibiotic as is evident from the fact that its genome contains the largest known resistance island with more than 50 resistance genes (Wikipedia). This is an "antibiotic paradox" because the very first antibiotic to be discovered in the modern world, was pyocyanase, discovered in 1888 by E. de Freudenreich. It was a mixture of several antibacterially active substances: pyocyanin, 1-hydroxyphenazine and an oily fraction. This natural antibiotic was able to kill typhoid, diphtheria and plague bacteria. Pyocyanase has been produced commercially, but unfortunately it was unstable and toxic. (pyocyanin and 1-hydroxyphenazine are also antifungal agents active against Candida albicans and Aspergillus fumigatus; Kerr J R et al., 1999). Pseudomonas aeruginosa colonies on blood agarPseudomonas aeruginosa on tryptic soy agarPseudomonas aeruginosa on MacConkey agarPseudomonas aeruginosa on Endo agarPseudomonas aeruginosa on cetrimide agarPseudomonas aeruginosa on various cultivation mediaPseudomonas aeruginosa identificationPseudomonas aeruginosa Gram stainPseudomonas aeruginosa SEMUseful LinksWIKIPEDIA CENTERS FOR DISEASE CONTROL AND PREVENTION (CDC) Todar's Online Textbook of Bacteriology Microbe Wiki (the student-edited microbiology resource) www.antimicrobe.org www.vetbact.org Merck Manualwww.microbiologyinpictures.com Pseudomonas aeruginosa - microbewikiA Microbial Biorealm page on the genus Pseudomonas aeruginosa Classification Higher order taxa
Species
 Description and significancePseudomonas aeruginosa is a gram-negative, rod-shaped, asporogenous, and monoflagellated bacterium that has an incredible nutritional versatility. It is a rod about 1-5 µm long and 0.5-1.0 µm wide. P. aeruginosa is an obligate respirer, using aerobic respiration (with oxygen) as its optimal metabolism although can also respire anaerobically on nitrate or other alternative electron acceptors. P. aeruginosa can catabolize a wide range of organic molecules, including organic compounds such as benzoate. This, then, makes P. aeruginosa a very ubiquitous microorganism, for it has been found in environments such as soil, water, humans, animals, plants, sewage, and hospitals (1). In all oligotropic aquatic ecosystems, which contain high-dissolved oxygen content but low plant nutrients throughout, P.aeruginosa is the predominant inhabitant and this clearly makes it the most abundant organism on earth (2). P.aeruginosa is an opportunistic human pathogen. It is “opportunistic” because it seldom infects healthy individuals. Instead, it often colonizes immunocompromised patients, like those with cystic fibrosis, cancer, or AIDS (3). It is such a potent pathogen that firstly, it attacks up two thirds of the critically-ill hospitalized patients, and this usually portends more invasive diseases. Secondly, P.aeruginosa is a leading Gram-negative opportunistic pathogen at most medical centers, carrying a 40-60% mortality rate. Thirdly, it complicates 90% of cystic fibrosis deaths; and lastly, it is always listed as one of the top three most frequent Gram-negative pathogens and is linked to the worst visual diseases (4). Furthermore, P.aeruginosa is a very important soil bacterium that is capable of breaking down polycyclic aromatic hydrocarbons and making rhamnolipids, quinolones, hydrogen cyanide, phenazines, and lectins (5). It also exhibits intrinsic resistance to a lot of different types of chemotherapeutic agents and antibiotics, making it a very hard pathogen to eliminate (1).  P. aeruginosa was first described as a distinct bacterial species at the end of the nineteenth century, after the development of sterile culture media by Pasteur. In 1882, the first scientific study on P. aeruginosa, entitled “On the blue and green coloration of bandages,” was published by a pharmacist named Carle Gessard. This study showed P. aeruginosa’s characteristic pigmentation: P. aeruginosa produced water-soluble pigments, which, on exposure to ultraviolet light, fluoresced blue-green light. This was later attributed to pyocyanine, a derivative of phenazine, and it also reflected the organism’s old names: Bacillus pyocyaneus, Bakterium aeruginosa, Pseudomonas polycolor, and Pseudomonas pyocyaneus (3). P. aeruginosa has many strains, including Pseudomonas aeruginosa strain PA01, Pseudomonas aeruginosa PA7, Pseudomonas aeruginosa strain UCBPP-PA14, and Pseudomonas aeruginosa strain 2192 (5). Most of these were isolated based on their distinctive grapelike odor of aminoacetophenone, pyocyanin production, and the colonies’ structure on agar media (6). Genome structureP. aeruginosa has the genome size of about 5.2 to 7 million base pairs (Mbp) with 65% Guanine + Cytosine content. It is a combination of variable accessory segments and a conserved core. The variable accessory genome is characterized by a set of genomic islands and islets from a primeval tRNA-integrated island type. The core genome consists of a low level of nucleotide divergence of 0.5% and a conserved synteny of genes, which means two or more genes, whether they are linked or not, are on the same chromosome (7).  P. aeruginosa has a single and supercoiled circular chromosome in the cytoplasm (4). It also carries a lot of chromosome-mobilizing plasmids that are very significant to the organism’s lifestyle as a pathogen. The plasmids, TEM, OXA, and PSE, for instance, are encoded for betalactamase production, which is necessary for its resistance to antibiotics, thus allowing P. aeruginosa to be a formidable pathogen (8). The two strains that have the complete genome sequence are Pseudomonas aeruginosa PA01 and Pseudomonas aeruginosa PA14 (9): --In 2000, a group of volunteer "Pseudomonas scientists", including those from the Washington PathoGenesis Corportaion and the Department of Biology of the University of California, San Diego, worked under the Pseudomonas aeruginosa Community Annotation Project (PseudoCAP) to publish the complete genome sequence of Pseudomonas aeruginosa PA01. This was done because knowing the genomic sequence would provide new information about this bacterium as a pathogen and about its ecological versatility and genetic complexity. At 6,264,403 base pairs, its bacterial genome is the largest to ever be sequenced. It also contains 5,570 predicted open reading frames (ORFs), and thus it almost has the genetic complexity of simple eukaryotes, such as Saccharomyces cerevisiae. Using whole-genome-shotgun sampling, the complete 6.3 Mbp genome of Pseudomonas aeruginosa PA01 is very much similar to the P. aeruginosa’s physical map, with only one major exception, which is the inversion of about a quarter of the Pseudomonas aeruginosa PA01 genome. This inversion comes from the homologous recombination of the rrnA and rrnB loci, and earlier studies on genomic sequence inversions of ribosomal DNA loci in S. typhimurium and E. coli suggest that this inversion might have adaptive significance (10). --The complete genome sequence of Pseudomonas aeruginosa PA14 is currently being done by Harvard Medical School scientists. The goal of this study is achieve a public data of Pseudomonas aeruginosa PA14 genome. The shotgun-sequencing phase of the project was finished in 2005, yielding 6.54 Mbp of PA14 sequence. It is currently being compared to the genome of Pseudomonas aeruginosa PA01 and preliminary results have shown that they are very similar but have several regions of marked differences, such as the insertion of the 107911bp in PA14, which is absent in PA01. Approximately, there is 96.3% of the DNA sequence of PAO1 is in PA14, and 92.4% of PA14 DNA sequence is in PA01 (11). Cell structure and metabolismProtein F--Since P. aeruginosa is a Gram-negative microbe, it has an outer membrane which contains Protein F (OprF). OprF functions as a porin, allowing certain molecules and ions to come into the cells, and as a structural protein, maintaining the bacterial cell shape. Because OprF provides P. aeruginosa outer membrane with an exclusion limit of 500 Da, it lowers the permeability of the outer membrane, a property that is desired because it would decrease the intake of harmful substances into the cell and give P. aeruginosa a high resistance to antibiotics (12). Flagellum and Pili--P. aeruginosa uses its single and polar flagellum to move around and to display chemotaxis to useful molecules, like sugars. Its strains either have a-type or b-type of flagella, a classification that is based primarily on the size and antigenicity of the flagellin subunit. The flagellum is very important during the early stages of infection, for it can attach to and invade tissues of the hosts (13). Similarly to its flagellum, P. aeruginosa pili contribute greatly to its ability to adhere to mucosal surfaces and epithelial cells. Specifically, it is the pili’s tip that is responsible for the adherence to the host cell surface. P. aeruginosa have N-methyl-phenyl-alanine (NMePhe) or type IV pili (1). The pili are characterized as long polar filaments made up of homopolymers from the protein pilin, which is encoded by the pilA gene (4). Overall, P. aeruginosa flagellum and pili have similar functionality (for attachment) and structure (both are filamentous structures on the surface of the cell), and their motility is controlled by RpoN, especially during initial attachment to the human host and under low nutrient conditions (1). 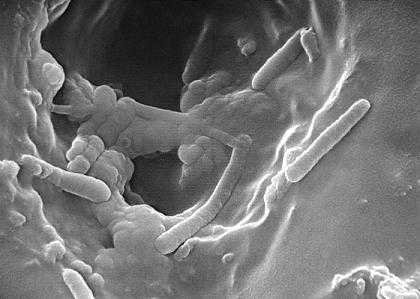 When infecting its host, P. aeruginosa is starved for iron because iron deprivation of an infecting pathogen is the key part in the humans’ innate defense mechanism. To overcome this challenge, P. aeruginosa synthesizes two siderophores: pyochelin and pyoverdin. P. aeruginosa then secrets these sideophores to the exterior of the cell, where they bind tightly to iron and bring the iron back into the cell. Additionally, P. aeruginosa can also use iron from enterobactin, a special siderophore produced by E. coli for iron transport, to satisfy its iron need (14). P. aeruginosa is a facultative aerobe; its preferred metabolism is respiration. It gains energy by transferring electrons from glucose, a reduced substrate, to oxygen, the final electron acceptor (15). The breakdown of glucose requires it to oxidize to gluconate in the periplasm, then it will be brought inside the inner membrane by a specific energy-dependent gluconate uptake system. Once inside, gluconate is phosphorylated to 6-P-gluconate, which will enter the central metabolism to produce energy for the cell (16). When P. aeruginosa is in anaerobic conditions, however, P. aeruginosa uses nitrate as a terminal electron acceptor(17). Under oxidative-stress conditions, P. aeruginosa synthesizes Fe- or Mn- containing superoxide dismutase (SOD) enzymes, which catalyze the very reactive O- to h3O2 and O2. It also detoxifies h3O2 to O2 and h3O by using catalase (1). EcologySince P. aeruginosa can live in both inanimate and human environments, it has been characterized as a “ubiquitous” microorganism. This versatility is made possible by a large number of enzymes that allow P. aeruginosa to use a diversity of substances as nutrients. Most impressively, P. aeruginosa can switch from growing on nonmucoid to mucoid environments, which comes with a large synthesis of alginate. In inanimate environment, P. aeruginosa is usually detected in water-reservoirs polluted by animals and humans, such as sewage and sinks inside and outside of hospitals. It is also found in swimming pools and whirlpools because the warm temperatures are favorable to its growth (3). Because it thrived in warm conditions, however, it was determined to be the culprit of the Hot Tub Rash, in which direct contact between the skin and the infected water from the tub will make the infected skin itchy and turn it a bumpy red color (19). In addition, P. aeruginosa is an opportunistic human pathogen that causes chronic infections in patients with cystic fibrosis and is the leading cause of death by Gram-negative bacteria (more under pathology) (3). Although most P. aeruginosa-plant interactions are detrimental to the plant, a recent study has found a P. aeruginosa strain that actually supports plant growth. This characteristic, along with the fact that P. aeruginosa can degrade polycyclic aromatic hydrocarbons, suggests the future uses of P. aeruginosa for environmental detoxification of synthetic chemicals and pesticides and for industrial purposes (3). Psuedomonas aeruginosa is unique due to its ability to infect both humans and plants, one of the few organisms that can infect both kingdoms.  P. aeruginosa groups tend to form biofilms, which are complex bacterial communities that adhere to a variety of surfaces, including metals, plastics, medical implant materials, and tissue. Biofilms are characterized by “attached for survival” because once they are formed, they are very difficult to destroy. Depending on their locations, biofilms can either be beneficial and detrimental to the environment. For instance, the biofilms found on rocks and pebbles underwater of lakes and ponds are an important food source for many aquatic organisms. On the contrary, those that developed on the interiors of water pipes might cause clogging and corrosions (19) (20). PathologyP. aeruginosa rarely causes disease in healthy humans. It is usually linked with patients whose immune system is compromised by diseases or trauma. It gains access to these patients’ tissues through the burns, for the burn victims, or through an underlying disease, like cystic fibrosis. First, P. aeruginosa adheres to tissue surfaces using its flagellum, pili, and exo-S; then, it replicates to create infectious critical mass; and lastly, it makes tissue damage using its virulence factors (21). Since the powerful exotoxins and endotoxins released by P. aeruginosa during bacteremias continue to infect the host even after P. aeruginosa has been killed off by antibiotics, acute diseases caused by P. aeruginosa tend to be chronic and life-threatening. Furthermore, with the exception of the cystic fibrosis strain, most P. aeruginosa strains that attack compromised patients tend to be nonmucoid (2). And even though a small amount of patients infected by P. aeruginosa developed severe sepsis with lesions with black centers, most patients exhibited no obvious pathological effects of the colonization (22). Cystic fibrosis (CF) is the most common autosomal recessive disorder in Caucasians. With a mutation on chromosome 7, a CF lung cannot transport chloride (Cl-), sodium (Na+), and water from the basolateral to the secretory epithelia. This disruption in the salt and water balance in the cell results in the production of a thick mucus, which becomes the ideal home for potential pathogens. P. aeruginosa attacks CF patients via airway and once it is in, it uses its flagellum to go to the hypoxic zone, an oxygen-depleted environment. At this location, P. aeruginosa undergoes a transition from an aerobic to an anaerobic microbe and starts forming biofilms anaerobically. Once this is formed, the P. aeruginosa in this community can sense their population via quorum sensing, where they secret low molecular weight pheromones that enable them to communicate with each other (23). This gives them the ability to resist many defenses, including anti-Pseudomonas antibiotics such as ticarcillin, ceftazidime, tobramycin, and ciprofloxacin, because once the bacteria sense that their outer layer of biofilm is being destroyed, the inner layers will grow stronger to reestablish the community (24). P. aeruginosa is also resistant to many antibiotics and chemotherapeutic agents due to their intrinsic resistance. This is caused by the low permeability to antibiotics of the outer membrane and by the production of β-lactamases against multidrug efflux pumps and β-lactam antibiotics (22). P. aeruginosa communicates with other cells through quorum-sensing. This form of communication allows the cells to regulate gene production which results in control of certain cell functions. One of the enzymes responsible for quorum sensing is tyrosine phosphatase (TpbA). This enzyme relays extracellular quorum sensing signals to polysaccharide production and biofilm formation outside the cells (32). P. aeruginosa attaches to surfaces by way of biofilm production. Quorum-sensing can be a drug target to cure infections caused by P. aeruginosa. Quorum-quenching is used to blocks the signaling mechanism of quorum-sensing and prevents biofilm formation in P. aeruginosa. Yi-Hu Dong and his colleagues were able to prevent biofilm formation in mice under laboratory conditions (33). P. aeruginosa secrets many virulent factors to colonize the cells of its host. For example, exotoxin A, the most toxic protein produced by P. aeruginosa, catalyzes the ADP-ribosylation to form ADP-ribosyl-EF-2, which inhibits the protein synthesis of the host’s cells. Moreover, elastase, an extracellular zinc protease, attacks eukaryotic proteins such as collagen and elastin and destroys the structural proteins of the cell. It also breaks down human immunoglobin and serum alpha proteins (1). Furthermore, P. aeruginosa infects animals. In an experiment, intravenous injection of virulent P. aeruginosa was injected into mice and these animals usually died within 24-48 hours. When a smaller dose was injected, characteristic signs of infection such as weight loss, focal lesions in liver, spleen, and kidneys, followed by death within 3-10 days, would take place. P. aeruginosa has also been found to cause outbreaks of pneumonia in guinea pigs, and although it also attacks plants, not a lot of research has been done in this area (22). Pseudomonas aeruginosa is an environmentally ubiquitous opportunistic pathogen. Epidermal infections often result from P. aeruginosa infiltrating through a human host’s first line of defenses, entering the body through the skin at the site of an open wound. P. aeruginosa is a common member of hospital bacterial communities where it can infect immunocompromised individuals including burn victims. P. aeruginosa is a source of bacteremia in burn victims [36]. Following severe skin damage, the prevalence of P. aeruginosa in the environment increases the probability of the organism accessing the bloodstream through the burn victim’s exposed deep epidermal tissue [36]. Previous research of antibody-mediated host defenses indicates that on the fifth day after the initial burn, Fc receptor expression is reduced in polymorphonuclear leukocytes (PMNs). Without the Fc receptor, PMN chemotaxis is greatly reduced and the PMNs become less effective at preventing infection [36]. P. aeruginosa can be transmitted to a host via fomites, vectors, and hospital workers who are potential carriers for multiply-antibiotic-resistant strains of the pathogen. Furthermore, any P. aeruginosa already present on a burn victim’s skin before the injury can transform from an innocuous organism on the surface of the skin to a source of infection in the bloodstream and body tissues of the same individual [36]. The pili and flagella of P. aeruginosa play a vital role in the infection of burns and wounds [36]. Controlled infection of burn wounds on animal and plant models with P. aeruginosa strains devoid of pili and flagella demonstrate a trend of decreased virulence. Without these morphological virulence factors, the bacteria exhibit a substantially decreased survival rate at the wound site and a decreased ability to disseminate within the host organism [36]. The spread of P. aeruginosa within host organisms is also dependent on the microorganism’s elastase production and other protease mechanisms. Bacterial elastase and other bacterial proteases degrade the host’s proteins, including the structural proteins within membranes, disrupting the host’s physical barriers against the spread of infection. Elastase also assists P. aeruginosa in avoiding phagocytotic antibody-mediated cytotoxicity at the site of the wound by inhibiting monocyte chemotaxis [36]. Identification:Macro morphology (smell): Large, flat and greenish colonies (2-4 mm in diameter) with irregular edges and typical metallic luster. The color is most visible on for instance TS-agar. Sometimes, a clear hemolysis zone is obtained on blood agar. Has distinctive smell (caramel, strawberry or raspberry soda). Some strains produce a green fluorescent pigment, pyoverdine. Some strains can also produce a blue pigment, pyocyanin.Micromorphology: Small motile rod (0.5-0.8 x 1.5-3 µm) with a monotrichous flagellum. Gram -: Fig. 65:5.Gram staining of Pseudomonas aeruginosa, strain ATCC 27853. The field B is a partial magnification (3 times) of A. The length of the scale bar corresponds to 5 µm. Date: 2011-03-24. G- Metabolism: Is often classified as aerobic, but can also exploit NO3- as final electron acceptor in the respiratory chain. Should, therefore, be classified as facultatively anaerobic! Catalase/Oxidase: +/+ Tryptophanase - Citrate +, methyl red -, Voges-Proskauer -. Spec. Char.: Temperature optimum: 37°C, but can also grow at 42°C. Hosts: Cattle, dog, horse, mink, poultry, sheep, reptiles etc. (including humans) Reservoir: The environment: soil, water etc. Disease (Swedish): Mastit, pneumoni, otit mm Disease (English): Mastitis, pneumonia, otitis, etc Application to BiotechnologyP. aeruginosa, as well as many other Pseudomonas, can degrade aromatic hydrocarbons such as methylbenzenes, which are the by-products of petroleum industries and are commonly used as solvents for enamels and paints as well as in the production of drugs and chemicals. Methylbenzenes are considered as environmental contaminants that are present in the atmosphere, underground and soils, and in surface water (25). P. aeruginosa can break down toluene, the simplest form of methylbenzene. P. aeruginosa degrades toluene through the oxidation of the methyl group to aldehyde, alcohol, and an acid, which is then converted to catechol. Hence, P. aeruginosa can be used in pollution control (26). Current ResearchEffect of Spaceflight on Microbial Gene Expression and Virulence (Microbe) --The National Aeronautics and Space Administration (NASA) and the Biodesign Institute at Arizona State University are currently carrying out a research project called the Microbe Experiment. In this experiment, three microbial pathogens Pseudomonas aeruginosa, Salmonella typhimurium, and Candida albicans are being brought into space to see how their genetic responses and virulence change. These three microbes have been viewed as potential threat to the health of the astronauts, for P. aeruginosa had contaminated the spacecraft’s water system and infected a crew member during the Apollo era. Thus, understanding their adaptation and virulence in microgravity will give scientists more information about the crew’s space environment and better prepare the astronauts for future space explorations. The microbes were placed inside self-contained culture chambers and upon landing back on earth, one thirds of the sample will be used for virulence studies while the remaining will be kept frozen at -80oC. Because this is an ongoing research project, there have not been any results but NASA scientists are very hopeful that this study will lead to novel discoveries of vaccines against these microbes here on Earth and during spaceflight (27). The Combination of PCR and Serology Increases the Diagnosis of Pseudomonas aeruginosa Colonization/Infection in Cystic Fibrosis --Microbiological culturing methods are often used for the early diagnosis of P. aeruginosa infection in cystic fibrosis (CF) patients. These methods, however, have some disadvantages because P. aeruginosa might not be detected since initial infection is usually in low density. It was then proposed that serology and polymerase chain reaction (PCR) might be better techniques in detecting the early stage of P. aeruginosa infection in children with CF. The experiment was carried out by collecting sputum and serum from 87 CF children with a mean age of 9.7 years. Then, 1) PCR was performed on the sputum, targeting P. aeruginosa algD GDP mannose dehydrogenase gene. 2) Serology was done against P. aeruginosa antigens: exotoxin A, elastase, and alkaline protease. 3) A combination of PCR and serology was done. When looking at the results, using the PCR or serology method alone did not yield statistically significant difference from the microbiological culturing methods. The combination of PCR and serology, however, identified a lot more patients than any of the two methods alone. Hence, a combination method that includes PCR will be an accurate technique to use in early diagnosis of P. aeruginosa colonization in CF patients (28). Genomic analysis reveals that Pseudomonas aeruginosa virulence is combinatorial --Comparison between the P. aeruginosa PA01 strain and the more virulent P. aeruginosa PA14 was done to identity new virulence genes. First, shotgun genome sequencing was done on PA14 using 65,800 plasmids with 2-4 kb fragments of PA14 DNA. Then, a long-range PCR based method was implemented to determine if certain P. aeruginosa genomes are similar to PA01 or PA14 genomes. It was found that although PA14 gemone (6.5 Mbp) is somewhat larger than that of PA01 (6.3 Mbp), PA14 and PA01 genomes are very similar. There were 58 gene clusters from PA14 that were missing in PA01 and it was assumed that some of these genes are what make PA14 a lot more virulent than PA01. Microarray genomotyping of 18 diverse strains in the C. elegans model, however, showed that those 58 PA14 gene clusters did not correlate with these strains’ virulence. Thus a conclusion was drawn that the virulence in P. aeruginosa is both combinatorial and multifactorial and that the genes required for one strain to be pathogenic are not required for virulence in other strains (29).  UV light test of fluorescent yellow/green pigment in nutrient broth Research also done in a microbiology lab at Loyola University Chicago has concluded that the Pseudomonas aeruginosa develops a greenish/yellow fluorescent pigment in nutrient broth and casein hydrolysis. After placing this fluorescent pigment under UV light, we observed a fluorescent blue/green pigment within the test tube. (35)  Fig. 6A Thr Under iron deficiency, a yellowish-green fluorescent pigment develops as a result of pyoverdins, a term named by Turfreijer for a group of compounds having a (1S)-5-amino-2,3-dihydro-8,9-dihydroxy-1H-pyrimido-[1,2a] chinolin-1-carboxylic acid chromophore. Figure 6A shows the different types of Pyoverdin groups that can be made on variations of their peptide chain. These pigment compounds only grow under iron limitation in a growth medium. Figure 6A shows the three different sv subgroups of Pyoverdin of P. aeruginosa. Since these three pyoverdin structures (also known as ferri-pyoverdins) are not produced by any other species of Pseudomonas they can be a quick way to identify the specific bacteria, Pseudomonas aeruginos. (34)References1) Lederberg, Joshua et al. Pseudomonas. Encyclopedia of Microbiology. Second Edition. Volume 3. San Diego, 2000. p. 876-891. 2) Costerton, W., and Anwar, H. Pseudomonas aeruginosa: The Microbe and Pathogen. Pseudomonas aeruginosa Infections and Treatment. 1994. p.1-17. 3) Botzenhardt, K., and Doring, G. Ecology and epidemiology of Pseudomonas aeruginosa. Pseudomonas aeruginosa as an Opportunistic Pathogen. 1993. p. 1-7. 4) Fick, R. Pseudomonas aeruginosa—the Microbial Hyena and Its Role in Disease: An Introduciton. Pseudomonas aeruginosa: The Opportunist. 1993. p. 1-6. 5) National Center for Biotechnology Information site 6) Gilardi, G. Cultural and Biochemical Aspects for Identification of Glucose-Nonfermenting Gram-Negative Rods. Nonfermenting Gram-Negative Rods. 1985. p.17-24. 7) Wiehlmann, L., Wagner, G., Cramer*, N., Siebert, B., Gudowius, P., Morales, G., Ko, T., Delden, C., Weinel, C., Slickers, P., and Tu, B. “Population structure of Pseudomonas aeruginosa”. Preceedings of the National Academy of Sciences of the United States of America. 2007. Volume 104. p. 8101–8106. 8) Craig, W., and Ebert, S. Antimirobial Therapy in Pseudomonas aeruginosa Infections. Pseudomonas aeruginosa Infections and Treatment. 1994. p. 470-491. 9) Pseudomonas Genome Database 10) Stover, K., Pham, Q., Erwin, L., Mizoguchi, D., Warrener, P., Hickey, J., Brinkman, L., Hufnagle, O., Kowalid, D., M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody*, S. N. Coulter, Folger, R., Kas, A., Larbig, K., Lim², R., Smith², K., Spencer², D., Wong², K.,Wu², Z., Paulsenk, J., Reizer, Z., Saier, H., Hancock³, R., Lory, S., and Olson, V. “Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen”. Nature. 2000. Volume 406. p. 961-964. 11) P. aeruginosa PA14 Genomic Sequencing Project 12) Stanisich, V., and Richmond, M. Gene Transfer in the Genus Pseudomonas. Genetics and Biochemistry of Pseudomonas. 1975. p. 170-175. 13) Delden, C. Virulence Fators in Pseudomonas aeruginosa. Pseudomonas. 2004. p. 1-7. 14) Cox, C. Iron and the Virulence of Pseudomonas aeruginosa. Pseudomonas aeruginosa: The Opportunist. 1993. p. 41-45. 15) Rabaey, K., and Verstraete, W. “Microbial Fuel Cells: novel biotechnology for enegy generation”. TrendS in Biotechnology. 2005. Volume 23. 16) Rojo, F., and Dinamarca, A. Catabolite Repression and Physiological Control. Pseudomonas. 2004. p. 365-366. 17) Valls, M., Cases, I., and Lorenzo, V. Transcription Mediated by rpoN-Dependent Promoters. Pseudomonas. 2004. p. 398-302. 18) Center for Diseases Control and Prevention 19) Center for Biofilm Engineering 20) Brown, M., and Simith, A. Antimicrobial Agents and Biofilms. Medical Implications of Biofilms. 2003. p. 36-38. 21) Irvin, Randall. Attachment and Colonization of Pseudomonas aerugionsa: Role of the Surface Structures. 'Pseudomonas aeruginosa as an Opportunistic Pathogen. 1993. p. 19-36. 22) Lowbury, E. Biological Importance of Pseudomonas aeruginosa: Medical Aspects. Genetics and Biochemistry of Pseudomonas. 1975. p. 37-43. 23) Holden, M., and Williams, P. “Quorum Sensing”. Encyclopedia of Life Sciences. 2001. 24) Smiley, A., and Hassett, D. Pseudomonas aeruginosa Biofilm Infections in Cystic Fibrosis. Biofilms, Infection, and Antimicrobial Therapy. 2006. p. 155-158. 25) Pieper, D., Stadler-Fritzche, K., Scholomann, M., and Knackmuss, H. Metabolism of 2-Chloro-4-Methylphenoxyacetate by Alcaligenes eutrophus JMP 134: Implications for the Degradation of Chloro- and Methyl-Substituted Aromatics via ortho Cleavage. Pseudomonas Molecular Biology and Biotechnology. 1992. p. 268-272. 26) Johnson, G., and Olsen, R. “Multiple Pathways for Toluene Degradation in Burkholderia sp. Strain JS150.” Applied and Environmental Microbiology. 1997. Volume 63. p. 4047-4052. [1] 27) The National Aeronautics and Space Administration 28) Filho, L,, Tateno, A., Martins, K., Chernishev, A., Garcia, D., Haug,M., Meisner, C., Rodrigues, C., and Do, G. “The Combination of PCR and Serology Increases the Diagnosis of Pseudomonas aeruginosa Colonization/Infection in Cystic Fibrosis.” Pediatric Pulmonology. 2007. Volume 42. p. 938–944. 29) Lee, D., Urbach, J., Wu, G., Liberati, N., Feinbaum, R., Miyata, S., Diggins, L., He, J., Saucier, M., Déziel, E., Friedman, L., Li, L., Grills, G., Montgomery, K., Kucherlapati, R., Rahme, L., and Ausubel, F. “Genomic analysis reveals that Pseudomonas aeruginosa virulence is Combinatorial.” Genome Biology. 2007. Volume 7. [2] 30) Edited by students: Vivek Brahmbhatt and Varun Garg of M Glogowski at Loyola University. http://web.ebscohost.com/ehost/pdfviewer/pdfviewer?vid=4&hid=104&sid=0029bc6b-e73c-4b8c-8882-42c0b9d6cedf%40sessionmgr112. 31) Malone, Jacob G., Tina Jaeger, and Christian Spangler. "YfiBNR Mediates Cyclic Di-GMP Dependent Small Colony Variant Formation and Persistence in Pseudomonas Aeruginosa." PLoS Pathogens 7.3 (2010): 1-17. Academic Search Premiere. Web. 24 Apr. 2010. 32) Uedal, A. & Wood, T.K. (2009). "Connecting quorum sensing, c-di-GMP, pel polysaccharide, and biofilm formation in pseudomonas aeruginosa through tyrosine phosphatase TpbA (PA3885)." Public Library of Science Pathogens. June 2009 Volume 5(6). 33) Dong, Y.H., Wang, L.-H., & Zhang, L.-H. (2007). "Quorum-quenching microbial infections: mechanisms and implications." Philosophical Transactions of the Royal Society B, 362(1483), 1201-1211. 34) [1]Herbert. "Siderophores of the Human Pathogenic Fluorescent Pseudomonads." Current Topics in Medicinal Chemistry 1.1 (2001): 1-6. Academic Search Premiere. Web. 27 Apr. 2010. 35) Edited by students: Safi Khan and Michelle Chua of M Glogowski at Loyola University. http://web.ebscohost.com/ehost/pdfviewer/pdfviewer?vid=5&hid=119&sid=4ad96fae-039a-487e-8c86-3de5c67d301f%40sessionmgr104 36) Edited by students: Katie Bates and Ashley Mores of M Glogowski at Loyola University. Lyczak, JB, Cannon, CL, Pier, GB. “Establishment of Pseudomonas aeruginosa infection: lessons from a versatile opportunist”. Microbes and Infection. 2000. Volume 2. Issue 9. p. 1051-1060. 37) "VetBact." VetBact. N.p., n.d. Web. 38) Walker, Travis S., Harsh Pal Bais, Eric Déziel, Herbert P. Schweizer, Laurence G. Rahme, Ray Fall, and Jorge M. Vivanco. "Pseudomonas Aeruginosa-Plant Root Interactions. Pathogenicity, Biofilm Formation, and Root Exudation." Plant Physiology. The American Society for Plant Biologists, n.d. Web. 11 May 2016. Written by Chelsea Dao, a student of Rachel Larsen Edited by KLB Edited by students: Vivek Brahmbhatt and Varun Garg / Michelle Chua and Safi Khan of Mary Glogowski at Loyola University, Irina Rojas and Aaron Beguelin at Hamilton College microbewiki.kenyon.edu PseudomonasTo search the entire book, enter a term or phrase in the form belowCustom Search Pseudomonas aeruginosa (page 1) (This chapter has 4 pages) © Kenneth Todar, PhD
Pseudomonas aeruginosa is member of the Gamma Proteobacteria class of Bacteria. It is a Gram-negative, aerobic rod belonging to the bacterial family Pseudomonadaceae. Since the revisionist taxonomy based on conserved macromolecules (e.g. 16S ribosomal RNA) the family includes only members of the genus Pseudomonas which are cleaved into eight groups. Pseudomonas aeruginosa is the type species of its group. which contains 12 other members. Like other members of the genus, Pseudomonas aeruginosa is a free-living bacterium, commonly found in soil and water. However, it occurs regularly on the surfaces of plants and occasionally on the surfaces of animals. Members of the genus are well known to plant microbiologists because they are one of the few groups of bacteria that are true pathogens of plants. In fact, Pseudomonas aeruginosa is occasionally a pathogen of plants. However, Pseudomonas aeruginosa has become increasingly recognized as an emerging opportunistic pathogen of clinical relevance. Several different epidemiological studies track its occurrence as a nosocomial pathogen and indicate that antibiotic resistance is increasing in clinical isolates. Pseudomonas aeruginosa is an opportunistic pathogen, meaning that it exploits some break in the host defenses to initiate an infection. In fact, Pseudomonas aeruginosa is the epitome of an opportunistic pathogen of humans. The bacterium almost never infects uncompromised tissues, yet there is hardly any tissue that it cannot infect if the tissue defenses are compromised in some manner. It causes urinary tract infections, respiratory system infections, dermatitis, soft tissue infections, bacteremia, bone and joint infections, gastrointestinal infections and a variety of systemic infections, particularly in patients with severe burns and in cancer and AIDS patients who are immunosuppressed. Pseudomonas aeruginosa infection is a serious problem in patients hospitalized with cancer, cystic fibrosis, and burns. The case fatality rate in these patients is near 50 percent. Pseudomonas aeruginosa is primarily a nosocomial pathogen. According to the CDC, the overall incidence of P. aeruginosa infections in U.S. hospitals averages about 0.4 percent (4 per 1000 discharges), and the bacterium is the fourth most commonly-isolated nosocomial pathogen accounting for 10.1 percent of all hospital-acquired infections. Characteristics Pseudomonas aeruginosa is a Gram-negative rod measuring 0.5 to 0.8 µm by 1.5 to 3.0 µm. Almost all strains are motile by means of a single polar flagellum. The bacterium is ubiquitous in soil and water, and on surfaces in contact with soil or water. Its metabolism is respiratory and never fermentative, but it will grow in the absence of O2 if NO3 is available as a respiratory electron acceptor. The typical Pseudomonas bacterium in nature might be found in a biofilm, attached to some surface or substrate, or in a planktonic form, as a unicellular organism, actively swimming by means of its flagellum. Pseudomonas is one of the most vigorous, fast-swimming bacteria seen in hay infusions and pond water samples. In its natural habitat Pseudomonas aeruginosa is not particularly distinctive as a pseudomonad, but it does have a combination of physiological traits that are noteworthy and may relate to its pathogenesis. • Pseudomonas aeruginosa has very simple nutritional requirements. It is often observed "growing in distilled water", which is evidence of its minimal nutritional needs. In the laboratory, the simplest medium for growth of Pseudomonas aeruginosa consists of acetate as a source of carbon and ammonium sulfate as a source of nitrogen. • P. aeruginosa possesses the metabolic versatility for which pseudomonads are so renowned. Organic growth factors are not required, and it can use more than seventy-five organic compounds for growth. • Its optimum temperature for growth is 37 degrees, and it is able to grow at temperatures as high as 42 degrees. • It is tolerant to a wide variety of physical conditions, including temperature. It is resistant to high concentrations of salts and dyes, weak antiseptics, and many commonly used antibiotics. • Pseudomonas aeruginosa has a predilection for growth in moist environments, which is probably a reflection of its natural existence in soil and water. These natural properties of the bacterium undoubtedly contribute to its ecological success as an opportunistic pathogen. They also help explain the ubiquitous nature of the organism and its prominence as a nosocomial pathogen. P. aeruginosa isolates may produce three colony types. Natural isolates from soil or water typically produce a small, rough colony. Clinical samples, in general, yield one or another of two smooth colony types. One type has a fried-egg appearance which is large, smooth, with flat edges and an elevated appearance. Another type, frequently obtained from respiratory and urinary tract secretions, has a mucoid appearance, which is attributed to the production of alginate slime. The smooth and mucoid colonies are presumed to play a role in colonization and virulence.
P. aeruginosa strains produce two types of soluble pigments, the fluorescent pigment pyoverdin and the blue pigment pyocyanin. The latter is produced abundantly in media of low-iron content and functions in iron metabolism in the bacterium. Pyocyanin (from "pyocyaneus") refers to "blue pus", which is a characteristic of suppurative infections caused by Pseudomonas aeruginosa.
www.textbookofbacteriology.net PseudomonasPseudomonas (page 4)© Kenneth Todar, PhD Table 2. Diseases caused by Pseudomonas aeruginosa Endocarditis. Pseudomonas aeruginosa infects heart valves of IV drug users and prosthetic heart valves. The organism establishes itself on the endocardium by direct invasion from the blood stream.Respiratory infections. Respiratory infections caused by Pseudomonas aeruginosa occur almost exclusively in individuals with a compromised lower respiratory tract or a compromised systemic defense mechanism. Primary pneumonia occurs in patients with chronic lung disease and congestive heart failure. Bacteremic pneumonia commonly occurs in neutropenic cancer patients undergoing chemotherapy. Lower respiratory tract colonization of cystic fibrosis patients by mucoid strains of Pseudomonas aeruginosa is common and difficult, if not impossible, to eradicate. Bacteremia and septicemia. Pseudomonas aeruginosa causes bacteremia primarily in immunocompromised patients. Predisposing conditions include hematologic malignancies, immunodeficiency relating to AIDS, neutropenia, diabetes mellitus, and severe burns. Most Pseudomonas bacteremia is acquired in hospitals and nursing homes. Pseudomonas accounts for about 25 percent of all hospital acquired Gram-negative bacteremias. Central nervous system infections. Pseudomonas aeruginosa causes meningitis and brain abscesses. The organism invades the CNS from a contiguous structure such as the inner ear or paranasal sinus, or is inoculated directly by means of head trauma, surgery or invasive diagnostic procedures, or spreads from a distant site of infection such as the urinary tract. Ear infections including external otitis. Pseudomonas aeruginosa is the predominant bacterial pathogen in some cases of external otitis, including "swimmer's ear". The bacterium is infrequently found in the normal ear, but often inhabits the external auditory canal in association with injury, maceration, inflammation, or simply wet and humid conditions. Eye infections. Pseudomonas aeruginosa can cause devastating infections in the human eye. It is one of the most common causes of bacterial keratitis, and has been isolated as the etiologic agent of neonatal ophthalmia. Pseudomonas can colonize the ocular epithelium by means of a fimbrial attachment to sialic acid receptors. If the defenses of the environment are compromised in any way, the bacterium can proliferate rapidly through the production of enzymes such as elastase, alkaline protease and exotoxin A, and cause a rapidly destructive infection that can lead to loss of the entire eye. Bone and joint infections. Pseudomonas infections of bones and joints result from direct inoculation of the bacteria or the hematogenous spread of the bacteria from other primary sites of infection. Blood-borne infections are most often seen in IV drug users and in conjunction with urinary tract or pelvic infections. Pseudomonas aeruginosa has a particular tropism for fibrocartilagenous joints of the axial skeleton. Pseudomonas aeruginosa causes chronic contiguous osteomyelitis, usually resulting from direct inoculation of bone and is the most common pathogen implicated in osteochondritis after puncture wounds of the foot. Urinary tract infections. Urinary tract infections (UTI) caused by Pseudomonas aeruginosa are usually hospital-acquired and related to urinary tract catheterization, instrumentation or surgery. Pseudomonas aeruginosa is the third leading cause of hospital-acquired UTIs, accounting for about 12 percent of all infections of this type. The bacterium appears to be among the most adherent of common urinary pathogens to the bladder uroepithelium. As in the case of E. coli, urinary tract infection can occur via an ascending or descending route. In addition, Pseudomonas can invade the bloodstream from the urinary tract, and this is the source of nearly 40 percent of Pseudomonas bacteremias. Gastrointestinal infections. Pseudomonas aeruginosa can produce disease in any part of the gastrointestinal tract from the oropharynx to the rectum. As in other forms of Pseudomonas disease, those involving the GI tract occur primarily in immunocompromised individuals. The organism has been implicated in perirectal infections, pediatric diarrhea, typical gastroenteritis, and necrotizing enterocolitis. The GI tract is also an important portal of entry in Pseudomonas septicemia and bacteremia. Skin and soft tissue infections, including wound infections, pyoderma and dermatitis. Pseudomonas aeruginosa can cause a variety of skin infections, both localized and diffuse. The common predisposing factors are breakdown of the integument which may result from burns, trauma or dermatitis; high moisture conditions such as those found in the ear of swimmers and the toe webs of athletes, hikers and combat troops, in the perineal region and under diapers of infants, and on the skin of whirlpool and hot tub users. Individuals with AIDS are easily infected. Pseudomonas has also been implicated in folliculitis and unmanageable forms of acne vulgaris. Host DefensesMost strains of P. aeruginosaare resistant to killing in serum alone, but the addition of polymorphonuclear leukocytes results in bacterial killing. Killing is most efficient in the presence of type-specific opsonizing antibodies, directed primarily at the antigenic determinants of LPS. This suggests that phagocytosis is an important defense and that opsonizing antibody is the principal functional antibody in protecting from P. aeruginosa infections. Once P. aeruginosa infection is established, other antibodies, such as antitoxin, may be important in controlling disease. The observation that patients with diminished antibody responses (caused by underlying disease or associated therapy) have more frequent and more serious P. aeruginosa infections underscores the importance of antibody-mediated immunity in controlling Pseudomonas infections. unfortunately, cystic fibrosis is the exception. Most cystic fibrosis patients have high levels of circulating antibodies to bacterial antigens, but are unable to clear P. aeruginosa efficiently from their lungs. Cell-mediated immunity does not seem to play a major role in resistance or defense against Pseudomonas infections. Epidemiology and Control of Pseudomonas aeruginosa Infections Pseudomonas aeruginosa is a common inhabitant of soil, water, and vegetation. It is found on the skin of some healthy persons and has been isolated from the throat (5 percent) and stool (3 percent) of nonhospitalized patients. In some studies, gastrointestinal carriage rates increased in hospitalized patients to 20 percent within 72 hours of admission. Within the hospital, P. aeruginosa finds numerous reservoirs: disinfectants, respiratory equipment, food, sinks, taps, toilets, showers and mops. Furthermore, it is constantly reintroduced into the hospital environment on fruits, plants, vegetables, as well by visitors and patients transferred from other facilities. Spread occurs from patient to patient on the hands of hospital personnel, by direct patient contact with contaminated reservoirs, and by the ingestion of contaminated foods and water. The spread of P. aeruginosa can best be controlled by observing proper isolation procedures, aseptic technique, and careful cleaning and monitoring of respirators, catheters, and other instruments. Topical therapy of burn wounds with antibacterial agents such as silver sulfadiazine, coupled with surgical debridement, dramatically reduces the incidence of P. aeruginosa sepsis in burn patients. Pseudomonas aeruginosa is frequently resistant to many commonly used antibiotics. Although many strains are susceptible to gentamicin, tobramycin, colistin, and fluoroquinolins, resistant forms have developed. The combination of gentamicin and carbenicillin is frequently used to treat severe Pseudomonas infections. Several types of vaccines are being tested, but none is currently available for general use. END OF CHAPTERwww.textbookofbacteriology.net Pseudomonas aeruginosa - Information and Epidemiology Services © Dennis Kunkel Microscopy Pseudomonas aeruginosa is a Gram negative bacteria that is commonly found in the environment e.g. soil, water and other moist locations What diseases are caused by Pseudomonas aeruginosa?Pseudomonas aeruginosa is an opportunistic pathogen. The bacteria takes advantage of an individual's weakened immune system to create an infection and this organism also produces tissue-damaging toxins. Pseudomonas aeruginosa causes urinary tract infections, respiratory system infections, dermatitis, soft tissue infections, bacteremia, bone and joint infections, gastrointestinal infections and a variety of systemic infections, particularly in patients with severe burns and in cancer and AIDS patients who are immunosuppressed. Who is more susceptible to infection from Pseudomonas aeruginosa?This bacterium is of particular concern to individuals with cystic fibrosis who are highly susceptible to pseudomonal lung infections. Pseudomonas aeruginosa is also of grave concern to cancer and burn patients as well as those people who are immunocompromised. The case fatality rate for individuals infected with Pseudomonas aeruginosa approaches 50 percent. Epidemiology of Pseudomonas aeruginosaPseudomonas aeruginosa is primarily a nosocomial pathogen. According to the CDC, the overall incidence of P. aeruginosa infections in US hospitals averages about 0.4 percent (4 per 1000 discharges), and the bacterium is the fourth most commonly-isolated nosocomial pathogen accounting for 10.1 percent of all hospital-acquired infections. Within the hospital, P. aeruginosa finds numerous reservoirs: disinfectants, respiratory equipment, food, sinks, taps, and mops. This organism is often reintroduced into the hospital environment on fruits, plants, vegetables, as well by visitors and patients transferred from other facilities. Spread occurs from patient to patient on the hands of hospital personnel, by direct patient contact with contaminated reservoirs, and by the ingestion of contaminated foods and water. Incubation PeriodUsually 24-72 hours. Diagnosis of Pseudomonas aeruginosaDiagnosis of P. aeruginosa infection depends upon isolation and laboratory identification of the bacterium. It grows well on most laboratory media and commonly is isolated on blood agar or eosin-methylthionine blue agar. It is identified on the basis of its Gram morphology, inability to ferment lactose, a positive oxidase reaction, its fruity odor, and its ability to grow at 42° C. Fluorescence under ultraviolet light is helpful in early identification of P. aeruginosa colonies and may also help identify its presence in wounds. Treatment of Pseudomonas aeruginosaPseudomonas aeruginosa is frequently resistant to many commonly used antibiotics. Although many strains are susceptible to gentamicin, tobramycin, colistin, and amikacin, resistant forms have developed. The combination of gentamicin and carbenicillin is frequently used to treat severe Pseudomonas infections. Several types of vaccines are being tested, but none is currently available for general use. * - Sometimes (mis) spelled Pseudomonas aeroginosa Contact EHA Consulting Group today for more information about how we can assist your company. www.ehagroup.com Pseudomonas Aeruginosa BakterisiPseudomonas aeruginosa, çoğu toprak ve suda bulunur. Glikozu oksidasyon yoluyla parçalayan fakat fermentasyon yapmayan bakterilerdir. Katalaz (+), insan patojeni, gram (-), sitrat (+), metil kırmızısı (-), Voges Proskauer(-), aerobik, polar flagellası (0,5*1,5-3 mikron boyutlarınada) ile hareket edebilen çubuk şekilli bakterilerdir. Ekseriye tek hücreler halinde görünürler, fakat bazen üreme esnasında birkaç hücre bitişik kalarak kısa zincirler teşkil ettikleri görülür. Genç kültürler, üzerinde büyüyebildiği ortamlarda genellikle mavi-yeşil bir pigment çıkarır. Kültür yaşlandıkça bu renkler kahverengine döner. Proteolitik ve lipolitik aktivite göstermektedirler. Aerobik olmaları nedeni ile gıdaların yüzeyinde hızlı gelişebilmeleri sonucu okside ürünler ve mukoz madde oluştururlar. Kendi gelişmeleri için gerekli gelişme faktörleri ve vitaminleri sentezleme yeteneğindedirler. Diğer Pseudomonadlar gibi Pseudomonas aeruginosa da piyosiyanin (mavi-yeşil), piyorubin (kırmızı-kahverengi) ve flöressein (yeşil-sarı ve flöresan) gibi pigmentler üretir. Flöresseini tüm Pseudomaslar oluşturablirken; piyosiyanini sadece Pseudomonas aeruginosa oluşturabilir. Pseudomonadların pigment üretimlerini arttırmak için Pseudomonas agar P-F gibi özel besiyerleri geliştirilmiştir. Hemoglobini tam olarak hemoliz ettiğinden kanlı agar besiyerindeki kolonilerin etrafında temiz ve berrak zon oluşturur. Pseudomonas aeruginosa in-vitro koşullarda, inci beyazı koloni görüntüsü ve üzümsü kokusuyla tanınır. Organizmanın kesin tanısı 42 °C de üreme yetisinin ve pigment üretiminin incelenmesiyle konur. Kurumaya dirençlilikleri zayıftır. Pseudomonas aeruginosa motorin ve karosen içinde büyüyebilir. Süt içindede iyi ürer, sütün pıhtılaşmasında ve çıkardığı pigmentten dolayı yeşil renk almasına neden olur. Bu özelliği sayesinde “hidrokarbon kullanan bakteriler (HUM)“ ünvanını kazanmıştır. HUM bakterileri mikrobiyal korozyona sebep olurlar. HUM bakterileri, hidrokarbon bazlı yakıtların üzerinde bazen yanlışlıkla “algae“ olarak adlandırılan, koyu yeşil ve jölemsi katmanlar oluştururlar. Biyokimyasal özellikleri Kanlı agarda hemoliz yaparlar. Kanlı agarda üreyen klinik izolatlar sıklıkla beta hemolitiktirler. Mueller-Hinton agarı yeşil renge çevirirler. Jelatin ve koagule plazmayı eriterek parçalarlar. Glikozu oksidatif yolla parçalayıp asit yaparlar. Laktaz ve sakkarozu kullanamazlar. Oksidaz (+) olmaları ile Enterebacteriaceae familyası üyelerinden ayrılırlar. Asetamini deamine ederek amonyak oluştururlar. Nişastayı etkilemezler. Katalaz ve sitrat reaksiyonları (+) tir. Indol ve Hidrojensülfür oluşturmazlar. L-arjinin dihidrolaz oluştururken, lisin dekarboksilaz(LDC) ve ornitin dekarboksilaz(ODC) oluşturmazlar. Metil kırmızısı ve Voges-Proskauer (-)’tir. Nitratı nitrite redükte ederler. KCN'ye dirençlidirler. P.a, P.fluorescens den ayrı olarak metilen mavisini ve prontosilin rengini giderir. İsmi Pseudomonas, Yunanca ¨sahte birim¨ demektir. Sahte manasına gelen ¨pseudo¨ ve birim manasına gelen ¨monas¨ kelimelerinden türetilmiştir. Mikrobiyolojinin ilk yıllarında bu isim bütün mikroorganizmaları tanımlamak içi kullanılıyordu. Aeruginosa ise Latince'de bakır pası manasına gelmektedir. Patojenliği P. aeruginosa özellikle, bağışıklık yetersizliği olan hastalarda solunum ve idrar yollarının, yanıkların ve açık yaraların fırsatçı patojenidir aynı zamanda kanda da enfeksiyonlar yapabilir. Nozokomial (hastane kaynaklı) enfeksiyonların onda biri P. aeruginosa sebebiyledir. Kistik fibroz hastaları P. aeruginosa enfeksiyonlarına özellikle yatkındırlar. P. aeruginosa kirli küvet ve jakuziler gibi su kalitesinin düşük olduğu durumlara maruz kalındığında dermatite sebep olabilir. Duyarlıkları P. aeruginosa piperacillin, imipenem, tobramycin, ciprofloxacin,polimiksin E(kolistin) gibi antibiyotiklerle tedavi edilebilir. Penisilin ve diğer Beta Laktam antibiyotiklerine karşı doğal olarak dayanıklıdır.
Kaynak: tr.wikipedia.org www.hangihastaliga.com Pseudomonas AeruginosaWhat is Pseudomonas Aeruginosa
Pseudomonas aeruginosa is a medically important human representatives of the genus Pseudomonas. This is approximately 2-4 mm long, non-spore-forming Gram-negative rods with polar flagellum. Pseudomonas aeruginosa can be attributed to simple culture media cultivate easily. On solid media show is mostly flat, great gray, often slimy-growing colonies, the surface often has a metallic sheen. Serve as a carbon source Pseudomonas aeruginosa organic compounds, is required for their utilization of molecular oxygen as an electron acceptor. The bacterial counts therefore to obligate aerobes. The optimum temperature is increasing at 36 ° C. Growth is also at 41 ° C, but not possible at 4 ° C. In liquid media, the seed grows to form a so-called scum predominantly at the surface. Most strains produce pigment under appropriate conditions. The main colors are yellow-green pyoverdin (fluorescein) and the bluish-green pyocyanin. Due to the formation of pigment was Pseudomonas aeruginosa earlier than the blue-green pus bacteria (Bacterium pyocyaneum) refers. The term aeruginosa derived from the Latin word aerugo (verdigris) from. Pseudomonas Aeruginosa Pathogenesis
Pseudomonas aeruginosa is due to its high resistance and low nutrient demands in the environment is widespread. There he is, especially in damp places, such as in soil and surface water, on plants and fruits as well as in the colon of healthy people to find. When he wet seed can also contaminate many areas and products in the home environment. This includes the plumbing, cleaning supplies, medicines, cosmetics and liquids for storage of contact lenses. But it can also survive in a dry medium for some time. In the hospital put next to the sanitation, contaminated IV solutions and blood products, primarily ventilators and nebulisers, humidifiers, and dialysis equipment and fluids important extracorporeal pathogen reservoir of the high resistance of Pseudomonas aeruginosa also requires that possess some disinfectant insufficient efficacy. At the hospital, the number of patients who are colonized with Pseudomonas aeruginosa, with the duration of the stay. The settlement takes place preferably on moist skin. In intensive care patients is also often colonized the upper respiratory tract. The mechanisms of pathogenicity are complex. Virulence factors are fimbriae that mediate adhesion to the cell surface, as well as various enzymes and exotoxins that the bacterium produces. Elastase, and proteases facilitate the invasion, where they are supported by the hemolysins (especially phospholipase C) that cause membrane damage in the tissue cells. Of exotoxin A, probably the most important virulence factor, exoenzyme S and is known to negatively affect the protein. Pseudomonas aeruginosa is the most common pathogens causing nosocomial infections. According to the Hospital Infection Surveillance System (KISS) in Germany caused approximately 10% of all hospital infections caused by Pseudomonas aeruginosa. In most cases these are pneumonia, skin and wound infections and infections of the urogenital tract. During sepsis, the proportion caused by Pseudomonas aeruginosa, about 3%. The Pseudomonas sepsis is burdened with the highest fatality rate among all forms of sepsis. Other diseases are ecthyma gangrenosum, meningitis, otitis and eye infections. Pseudomonas aeruginosa infections occur mainly in patients with immune deficiency. Most cases are therefore found in the critical care area, in burn units and hematology-oncology at stations. Drug addicts are also dar. a risk group outside the hospital Pseudomonas aeruginosa plays as a pathogen only in patients with cystic fibrosis, bronchiectasis and a greater role in urological infections. Pseudomonas Aeruginosa Diagnosis
The microbiological diagnosis involves the isolation of the pathogen from the respective test materials and biochemical identification. Pseudomonas aeruginosa is naturally resistant to many antibiotics. Those available in Germany only the beta-lactam antibiotics Acylureidopenicilline [piperacillin (PIPRIL) + beta-lactamase inhibitor], aztreonam (AZACTAM), and certain cephalosporins [ceftazidime (Fortum), cefepime (Maxipime)] and carbapenems [imipenem (ZIENAM etc.) meropenem (Meron),] sufficiently effective. A good activity also show the aminoglycosides [amikacin (BIKLIN), gentamicin (Refobacin etc.), netilmicin (CERTOMYCIN), tobramycin (GERNEBCIN)] and fluoroquinolones [(ciprofloxacin (Cipro, etc.), levofloxacin (Tavanic)]. According to the study resistance of the Paul Ehrlich Society for Chemotherapy in 2004 in each case > 90% of the strains were sensitive to meropenem and tobramycin. For amikacin, cefepime, ceftazidime, ciprofloxacin and piperacillin / tazobactam (Tazobac) varied the proportion of sensitive strains between 75% and 80%. It should be noted, however, that the isolates from patients on general wards usually sensitive to antibiotics were significantly more likely than those in intensive care units. Pseudomonas Aeruginosa TherapyThe calculated treatment of infections where Pseudomonas aeruginosa is suspected as the causative agent, to keep up with the local resistance situation. To treat life-threatening infections are piperacillin / tazobactam, ceftazidime, cefepime, imipenem or meropenem, usually in combination with a Pseudomonas effective aminoglycoside or fluoroquinolone considered. For less severe infections may also be monotherapy with a beta-lactam antibiotic ciprofloxacin or be successful if adequate doses by the development of a resistant subpopulation is prevented. The treatment of Pseudomonas infections in patients with cystic fibrosis based on the antibiogram. If necessary. Macrolides are also due to their immunomodulatory properties for the treatment in question. For the initial treatment of malignant external otitis is piperacillin (12-20 g daily) in combination with tobramycin (0.24 g daily) for four weeks and then oral ciprofloxacin (g daily 1-1.5) over several months recommended.
pseudomonas-aeruginosa.org |
г.Самара, ул. Димитрова 131 [email protected] |
 |

 Gram stain of Pseudomonas aeruginosa cells
Gram stain of Pseudomonas aeruginosa cells
 Pseudomonas aeruginosa
colonies on agar
Pseudomonas aeruginosa
colonies on agar
 The
soluble blue pigment pyocyanin
is produced by many,
but not all, strains of Pseudomonas aeruginosa
The
soluble blue pigment pyocyanin
is produced by many,
but not all, strains of Pseudomonas aeruginosa